LiveScience | NASA's human spaceflight program might take some giant leaps forward if the agency embraces genetic engineering techniques more fully, according to genomics pioneer J. Craig Venter.
The biologist, who established the J. Craig Venter Institute that created the world's first synthetic organism earlier this year, told a crowd here Saturday (Oct. 30) that human space exploration could benefit from more genetic screening and genetic engineering. Such efforts could help better identify individuals most suited for long space missions, as well as make space travel safer and more efficient, he said.
"I think this could change the shape of what NASA does, if you make the commitment to do it," said Venter, who led a team that decoded the human genome a decade ago. Venter spoke to a group of scientists and engineers who gathered at NASA's Ames Research Center for two different meetings: a synthetic biology workshop put on by NASA, and Space Manufacturing 14: Critical Technologies for Space Settlement, organized by the nonprofit Space Studies Institute.
Astronauts with the right (genetic) stuff
Genetics techniques could come in extremely handy during NASA's astronaut selection process, Venter said. The space agency could screen candidates for certain genes that help make good spaceflyers — once those genes are identified, he added.
Genes that encode robust bone regeneration, for example, would be a plus, helping astronauts on long spaceflights battle the bone loss that is typically a major side effect of living in microgravity. Also a plus for any prospective astronaut: genes that code for rapid repair of DNA, which can be damaged by the high radiation levels in space.
Genetic screening would be a natural extension of what NASA already does — it would just add a level of precision, according to Venter.
"NASA's been doing genetic selection for a long time," he said. "You just don't call it that."
Last summer, the agency chose just nine astronaut candidates — out of a pool of 3,500 — for its rigorous astronaut training program based on a series of established spaceflight requirements and in-depth interviews.
A new microbiome
At some point down the road, NASA could also take advantage of genetic engineering techniques to make long space journeys more efficient and easier on astronauts, Venter said.
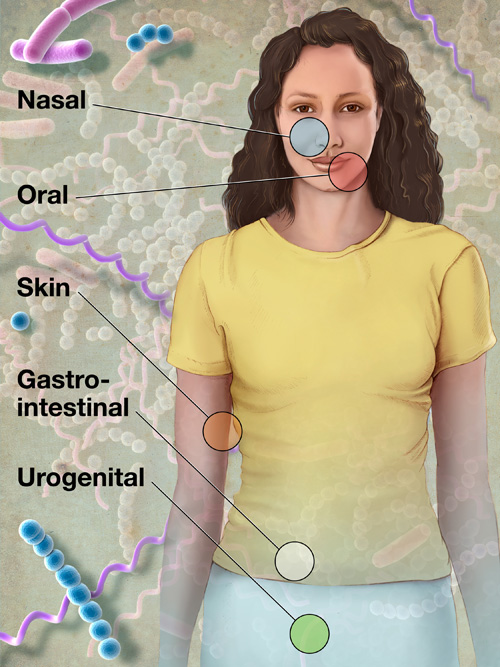
As an example, he cited the human microbiome, the teeming mass of microbes that live on and inside every one of us. Every human body hosts about 100 trillion microbes — meaning the bugs outnumber our own cells by a factor of at least 10 to one.
While humans only have about 20,000 genes, our microbiome boasts a collective 10 million or so, Venter said. These microbes provide a lot of services, from helping us digest our food to keeping our immune system's inflammation response from going overboard.
With some tailoring, the microbiome could help us out even more, according to Venter.
"Why not come up with a synthetic microbiome?" he asked.
Theoretically, scientists could engineer gut microbes that help astronauts take up nutrients more efficiently. A synthetic microbiome could also eliminate some pathogens, such as certain bacteria that can cause dental disease. Other tweaks could improve astronauts' living conditions, and perhaps their ability to get along with each other in close quarters.
Body odor is primarily caused by microbes, Venter said. A synthetic microbiome could get rid of the offenders, as well as many gut microbes responsible for excessive sulfur or methane production. Fist tap Nana.