Friday, July 19, 2013
of course cancer is caused by bacteria..., told you so years ago
By
CNu
at
July 19, 2013
2
comments
![]()
Labels: as above-so below , microcosmos
isn't smegma believed to cause cervical cancer?
By
CNu
at
July 19, 2013
0
comments
![]()
Labels: microcosmos
the doe has a "joint genome institute" exploring uncharted reaches of the microcosmos...,
By
CNu
at
July 19, 2013
0
comments
![]()
Labels: ecosystems , microcosmos , Possibilities , tactical evolution
pandoraviruses hint at fourth domain of life...,
 ,
have been dubbed "Pandoraviruses" because of the surprises they may
hold for biologists, in reference to the mythical Greek figure who
opened a box and released evil into the world.
,
have been dubbed "Pandoraviruses" because of the surprises they may
hold for biologists, in reference to the mythical Greek figure who
opened a box and released evil into the world.
By
CNu
at
July 19, 2013
0
comments
![]()
Labels: microcosmos
Sunday, April 07, 2013
human breath analysis and individual metabolic phenotypes
By
CNu
at
April 07, 2013
23
comments
![]()
Labels: microcosmos , symbiosis , What IT DO Shawty...
Thursday, March 28, 2013
energetic costs of cellular computations
By
CNu
at
March 28, 2013
0
comments
![]()
Labels: agency , microcosmos , What IT DO Shawty...
Saturday, March 09, 2013
lake vostok yields new bacterial life
By
CNu
at
March 09, 2013
3
comments
![]()
Labels: as above-so below , microcosmos
Saturday, November 24, 2012
afterlife...,
By
CNu
at
November 24, 2012
7
comments
![]()
Labels: microcosmos
Sunday, September 09, 2012
Monday, August 27, 2012
"told you so" moments stacking up like hotcakes now...,
 NYTimes | IN recent years, scientists have made extraordinary advances in understanding the causes of autism, now estimated to afflict 1 in 88 children. But remarkably little of this understanding has percolated into popular awareness, which often remains fixated on vaccines.
NYTimes | IN recent years, scientists have made extraordinary advances in understanding the causes of autism, now estimated to afflict 1 in 88 children. But remarkably little of this understanding has percolated into popular awareness, which often remains fixated on vaccines. So here’s the short of it: At least a subset of autism — perhaps one-third, and very likely more — looks like a type of inflammatory disease. And it begins in the womb.
It starts with what scientists call immune dysregulation. Ideally, your immune system should operate like an enlightened action hero, meting out inflammation precisely, accurately and with deadly force when necessary, but then quickly returning to a Zen-like calm. Doing so requires an optimal balance of pro- and anti-inflammatory muscle.
In autistic individuals, the immune system fails at this balancing act. Inflammatory signals dominate. Anti-inflammatory ones are inadequate. A state of chronic activation prevails. And the more skewed toward inflammation, the more acute the autistic symptoms.
Nowhere are the consequences of this dysregulation more evident than in the autistic brain. Spidery cells that help maintain neurons — called astroglia and microglia — are enlarged from chronic activation. Pro-inflammatory signaling molecules abound. Genes involved in inflammation are switched on.
These findings are important for many reasons, but perhaps the most noteworthy is that they provide evidence of an abnormal, continuing biological process. That means that there is finally a therapeutic target for a disorder defined by behavioral criteria like social impairments, difficulty communicating and repetitive behaviors.
But how to address it, and where to begin? That question has led scientists to the womb. A population-wide study from Denmark spanning two decades of births indicates that infection during pregnancy increases the risk of autism in the child. Hospitalization for a viral infection, like the flu, during the first trimester of pregnancy triples the odds. Bacterial infection, including of the urinary tract, during the second trimester increases chances by 40 percent.
The lesson here isn’t necessarily that viruses and bacteria directly damage the fetus. Rather, the mother’s attempt to repel invaders — her inflammatory response — seems at fault. Research by Paul Patterson, an expert in neuroimmunity at Caltech, demonstrates this important principle. Inflaming pregnant mice artificially — without a living infective agent — prompts behavioral problems in the young. In this model, autism results from collateral damage. It’s an unintended consequence of self-defense during pregnancy.
Yet to blame infections for the autism epidemic is folly. First, in the broadest sense, the epidemiology doesn’t jibe. Leo Kanner first described infantile autism in 1943. Diagnoses have increased tenfold, although a careful assessment suggests that the true increase in incidences is less than half that. But in that same period, viral and bacterial infections have generally declined. By many measures, we’re more infection-free than ever before in human history.
Better clues to the causes of the autism phenomenon come from parallel “epidemics.” The prevalence of inflammatory diseases in general has increased significantly in the past 60 years. As a group, they include asthma, now estimated to affect 1 in 10 children — at least double the prevalence of 1980 — and autoimmune disorders, which afflict 1 in 20.
By
CNu
at
August 27, 2012
0
comments
![]()
Labels: microcosmos , What IT DO Shawty...
Tuesday, August 21, 2012
but then I told you this a looooong time ago, right?
“They’re not exactly your flagship disease-causing bacteria,” lead researcher Christian Jobin, from the University of North Carolina at Chapel Hill, told Nature.
Individuals with inflammatory bowel disease are at higher risk of developing colorectal cancer than the general population. But researchers thought that the main culprit was the over-active immune cells, which released DNA-damaging molecules. The new work suggests that gut microbes may also contribute to the process, as inflammation appears to change the microbial composition of the gut to favor toxin-producing E. coli strains.
Experts think that the research could lead to methods of reducing the risk of cancer by altering the microbial community, though that strategy has to be tested.
By
CNu
at
August 21, 2012
0
comments
![]()
Labels: as above-so below , microcosmos , What IT DO Shawty...
Friday, June 22, 2012
medical ecology
“I would like to lose the language of warfare,” said Julie Segre, a senior investigator at the National Human Genome Research Institute. “It does a disservice to all the bacteria that have co-evolved with us and are maintaining the health of our bodies.”
This new approach to health is known as medical ecology. Rather than conducting indiscriminate slaughter, Dr. Segre and like-minded scientists want to be microbial wildlife managers.
No one wants to abandon antibiotics outright. But by nurturing the invisible ecosystem in and on our bodies, doctors may be able to find other ways to fight infectious diseases, and with less harmful side effects. Tending the microbiome may also help in the treatment of disorders that may not seem to have anything to do with bacteria, including obesity and diabetes.
“I cannot wait for this to become a big area of science,” said Michael A. Fischbach, a microbiologist at the University of California, San Francisco, and an author of a medical ecology manifesto published this month in the journal Science Translational Medicine.
Judging from a flood of recent findings about our inner ecosystem, that appears to be happening. Last week, Dr. Segre and about 200 other scientists published the most ambitious survey of the human microbiome yet. Known as the Human Microbiome Project, it is based on examinations of 242 healthy people tracked over two years. The scientists sequenced the genetic material of bacteria recovered from 15 or more sites on their subjects’ bodies, recovering more than five million genes.
The project and other studies like it are revealing some of the ways in which our invisible residents shape our lives, from birth to death.
A number of recent reports shed light on how mothers promote the health of their children by shaping their microbiomes. In a study published last week in the journal PLoS One, Dr. Kjersti Aagaard-Tillery, an obstetrician at Baylor College of Medicine, and her colleagues described the vaginal microbiome in pregnant women. Before she started the study, Dr. Aagaard-Tillery expected this microbiome to be no different from that of women who weren’t pregnant.
“In fact, what we found is the exact opposite,” she said.
By
CNu
at
June 22, 2012
5
comments
![]()
Labels: microcosmos , What IT DO Shawty...
Monday, June 18, 2012
oops, they forgot the breastesses...,
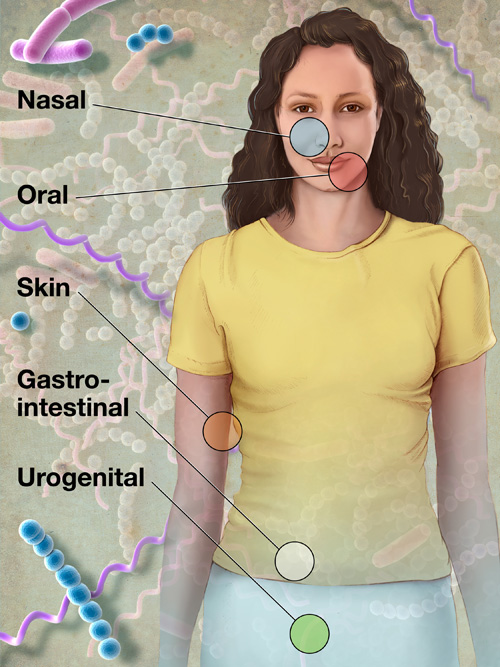 kurzweilai | Some 200 members of the Human Microbiome Project (HMP) Consortium from nearly 80 universities and scientific institutions, organized by the National Institutes of Health, have mapped the normal microbial makeup of healthy humans, producing numerous insights and even a few surprises.
kurzweilai | Some 200 members of the Human Microbiome Project (HMP) Consortium from nearly 80 universities and scientific institutions, organized by the National Institutes of Health, have mapped the normal microbial makeup of healthy humans, producing numerous insights and even a few surprises.The report on on their five years of research was published Thusday June 14, 2012, in a series of coordinated scientific reports in Nature the PLoS.
Researchers found, for example, that nearly everyone routinely carries pathogens, microorganisms known to cause illnesses.
In healthy individuals, however, pathogens cause no disease; they simply coexist with their host and the rest of the human microbiome, the collection of all microorganisms living in the human body.
Researchers must now figure out why some pathogens turn deadly and under what conditions, likely revising current concepts of how microorganisms cause disease.
“Like 15th century explorers describing the outline of a new continent, HMP researchers employed a new technological strategy to define, for the first time, the normal microbial makeup of the human body,” said NIH Director Francis S. Collins, M.D., Ph.D.
“HMP created a remarkable reference database by using genome sequencing techniques to detect microbes in healthy volunteers. This lays the foundation for accelerating infectious disease research previously impossible without this community resource.”
To define the normal human microbiome, HMP researchers sampled 242 healthy U.S. volunteers (129 male, 113 female), collecting tissues from 15 body sites in men and 18 body sites in women.
Researchers collected up to three samples from each volunteer at sites such as the mouth, nose, skin (two behind each ear and each inner elbow), and lower intestine (stool), and three vaginal sites in women; each body site can be inhabited by organisms as different as those in the Amazon Rainforest and the Sahara Desert.
Historically, doctors studied microorganisms in their patients by isolating pathogens and growing them in culture. This painstaking process typically identifies only a few microbial species, as they are hard to grow in the laboratory. In HMP, researchers purified all human and microbial DNA in each of more than 5,000 samples and analyzed them with DNA sequencing machines.
By
CNu
at
June 18, 2012
0
comments
![]()
Labels: microcosmos , What IT DO Shawty...
the wonder of breasts
 Guardian | We love breasts, yet can't quite take them seriously. Breasts embarrass us. They're unpredictable. They're goofy. They can turn babies and grown men into lunkheads.
Guardian | We love breasts, yet can't quite take them seriously. Breasts embarrass us. They're unpredictable. They're goofy. They can turn babies and grown men into lunkheads.They appear out of nowhere in puberty, they get bigger in pregnancy, they're capable of producing prodigious amounts of milk, and sometimes they get sick. But for such an enormously popular feature of the human race, it's remarkable how little we know about their basic biology.
The urgency to know and understand breasts has never been greater. Modern life has helped many of us live longer and more comfortably. It has also, however, taken a strange toll on our breasts. For one thing, they are bigger than ever. We are sprouting them at younger ages. We are filling them with saline and silicone and transplanted stem cells to change their shape. This year marks the 50th anniversary of the first silicone implant surgery in Houston, Texas.
More tumours form in the breast than in any other organ, making breast cancer the most common malignancy in women worldwide. Its incidence has almost doubled since the 1940s and is still rising.
But breasts are often overlooked, at least for non-cancer scientific research. The Human Microbiome Project, for example, is decoding the microbial genes of every major human gland, liquid and orifice, from the ears to the genitals. It neglected to include breast milk.
I wanted to know more, so I went to the 15th meeting of the International Society for Research in Human Milk and Lactation in Lima. Many attendees were molecular biologists, biochemists or geneticists who are deconstructing milk bit by bit. Until recently, it was thought breast milk had around 200 components. These could be divided into the major ingredients of fats, sugars, proteins and enzymes. But new technologies have allowed researchers to look deeper into each of these categories and discover new ones.
Scientists used to think breast milk was sterile, like urine. But it's more like cultured yoghurt, with lots of live bacteria doing who knows what. These organisms evolved for a reason, and somehow they're helping us out. One leading theory is they act as a vaccine, inoculating the infant gut. A milk sample has anywhere from one to 600 species of bacteria. Most are new to science.
Then there are the sugars. There's a class of them called oligosaccharides, which are long chains of complex sugars. Scientists have identified 140 of them so far, and estimate there are about 200. The human body is full of oligosaccharides, which ride on our cells attached to proteins and lipids. But a mother's mammary gland cooks up a unique batch of "free" or unattached ones and deposits them in milk. These are found nowhere else in nature, and not every mother produces the same ones, since they vary by blood type. Even though they're sugars, the oligosaccharides are, weirdly, not digestible by infants. Yet they are a main ingredient, present in milk in the same percentage as the proteins, and in higher amounts than the fats. So what are they doing there?
They don't feed us, but they do feed many types of beneficial bacteria that make a home in our guts and help us fight infections. In addition to recruiting the good bugs, these sugars prevent the bad bugs from hanging around. "The benefits of human milk are still underestimated," said Lars Bode, an immunobiologist at the University of California, San Diego. "We're still discovering functional components of breast milk."
By
CNu
at
June 18, 2012
1 comments
![]()
Labels: microcosmos , What IT DO Shawty...
Friday, May 18, 2012
what bugs are in your gut?
“It’s a humungous paper, with multiple key findings,” said food scientist David Mills of the University of California, Davis. “An impressive and complex piece of work,” agreed molecular biologist Jeremy Nicholson of Imperial College, London. Neither researcher participated in the study.
The scale and complexity stem from the research team’s aim of answering a multifaceted question—“What is the degree to which these microbial communities… vary within a person, as a function of postnatal development, physiological status, cultural tradition, and where a person lives,” said geneticist Jeffrey Gordon of the Washington University in St Louis, who led the study.
To this end, the researchers collected samples of feces from villagers in rural Malawi, Amerindians in Amazonian Venezuela, and metropolis-dwelling Americans. They then performed high-throughput sequencing on DNA taken from the samples to determine both the species and strains of microbes present and which microbial genes were most abundant.
The team found a common pattern for how the microbiomes of babies develop in the three countries. “It takes 6 to 9 months to get the first 6 or 700 bugs and then another couple of years to get the adult set,” explained Nicholson. “[Gordon] finds there is the same sort of developmental time span between countries,” he said, “but that the resulting microbiomes are nonetheless distinct between, let’s call it, a third-world population and a westernized population.”
One of the most striking differences was the degree of microbial diversity, with both the Amerindians and Malawians having far greater diversity than the Americans. “But, ironically, [Americans] might have more diversity in terms of the food eaten,” said Mills, which might have been expected to correlate with microbial diversity. Gordon suggested the Westerners’ lack of diversity could result from “our lifestyle, our degree of hygiene, [and] our use of antibiotics,” though further research is needed to test these possibilities.
Despite these differences between the gut microbiomes of the three cultures, there were also striking similarities, said Gordon. For example, “across all three populations, we see this age-dependent change in vitamin biosynthesis,” he said. In infants, gut bacteria tend to carry more copies of genes involved in folate biosynthesis, while the guts of older individuals harbor microbes carrying more genes for folate metabolism. Conversely, genes involved in vitamin B-12 synthesis became more prevalent in the gut microbiome with age.
“What’s really fascinating about those results,” said Mills, “is that it is reflecting what the host needs.”
By
CNu
at
May 18, 2012
0
comments
![]()
Labels: microcosmos , What IT DO Shawty...
Thursday, May 10, 2012
oh, it's more than that...., but at least this is a start
Of the 7.5m global deaths from cancer in 2008, an estimated 1.5m may have been due to potentially preventable or treatable infections.
Scientists carried out a statistical analysis of cancer incidence to calculate that around 16% of all cancers diagnosed in 2008 were infection-related. The proportion of cancers linked to infection was three times higher in developing countries than in developed ones.
Key cancer-causing infectious agents include human papillomavirus (HPV), the gastric bug Helicobacter pylori and the hepatitis B (HBV) and C viruses.
These four were together believed to be responsible for 1.9m cases of cancer, mostly gastric, liver and cervical cancers.
Cervical cancer accounted for around half of infection-related women's cancers. In men, more than 80% of infection-related cancers affected the liver, stomach and colon.
Dr Catherine de Martel and Dr Martyn Plummer, from the International Agency for Research on Cancer in Lyon, wrote in the Lancet Oncology journal: "Infections with certain viruses, bacteria, and parasites are one of the biggest and preventable causes of cancer worldwide … Application of existing public-health methods for infection prevention, such as vaccination, safer injection practice, or antimicrobial treatments, could have a substantial effect on future burden of cancer worldwide."
The researchers used information from a number of sources, including a cancer-incidence database covering 27 cancers from 184 countries.
By
CNu
at
May 10, 2012
0
comments
![]()
Labels: microcosmos
Tuesday, November 29, 2011
a truly great loss indeed...,
“She was always stimulating; she always had a new idea, some new connection she had seen and she couldn’t wait to tell you about,” Steve Goodwin, Dean of the College of Natural Resources and the Environment told MassLive.com.
Margulis showed early aptitude in science, enrolling at the University of Chicago and earning her bachelor’s degree in zoology by the age of 18. Shortly thereafter she married her first husband, the astronomer Carl Sagan. The marriage ended by the time she got her doctorate in genetics from the University of California, Berkeley, in 1965.
She developed her ideas on symbiosis in the late 1960s, and tried to publish her ideas in 15 journals before finally being accepted by the Journal of Theoretical Biology, according to The New York Times. Though it was highly controversial at the time, serial symbiosis is widely accepted among evolutionary scientists today.
In the 1970s, she became a supporter of James Lovelock’s Gaia hypothesis, which proposed that the earth could be thought of as a complex system whose atmospheric and mineral components existed in symbiosis with living organisms, allowing biota as a whole to self-perpetuate.
She taught evolutionary biology for nearly 40 years, first at the Boston University and then at the University of Massachusetts, where I had the opportunity to experience her carefully crafted course. I came to the class expecting Margulis to expound on the theories that she had championed. Instead, she exposed our small seminar class to the experiments of many researchers whose work provided evidence for her ideas, and invited us to make own conclusions.
“If science doesn’t fit in with the cultural milieu, people dismiss science—they never reject their cultural milieu!” said Margulis in the book The Third Culture: Beyond the Scientific Revolution. In the same chapter, Richard Dawkins wrote: “I greatly admire Lynn Margulis’s sheer courage and stamina in sticking by the endosymbiosis theory, and carrying it through from being an unorthodoxy to an orthodoxy.”
According to The New York Times, Margulis died from a stroke. She is survived by a daughter Jennifer Margulis and three sons Dorion Sagan, Jeremy Sagan, Zachary Margulis-Ohnuma.
By
CNu
at
November 29, 2011
1 comments
![]()
Labels: microcosmos , point source
Tuesday, October 18, 2011
what is nanotechnology?
 crnano | A basic definition: Nanotechnology is the engineering of functional systems at the molecular scale. This covers both current work and concepts that are more advanced.
crnano | A basic definition: Nanotechnology is the engineering of functional systems at the molecular scale. This covers both current work and concepts that are more advanced.In its original sense, 'nanotechnology' refers to the projected ability to construct items from the bottom up, using techniques and tools being developed today to make complete, high performance products.
The Meaning of Nanotechnology
When K. Eric Drexler popularized the word 'nanotechnology' in the 1980's, he was talking about building machines on the scale of molecules, a few nanometers wide—motors, robot arms, and even whole computers, far smaller than a cell. Drexler spent the next ten years describing and analyzing these incredible devices, and responding to accusations of science fiction. Meanwhile, mundane technology was developing the ability to build simple structures on a molecular scale. As nanotechnology became an accepted concept, the meaning of the word shifted to encompass the simpler kinds of nanometer-scale technology. The U.S. National Nanotechnology Initiative was created to fund this kind of nanotech: their definition includes anything smaller than 100 nanometers with novel properties.
Much of the work being done today that carries the name 'nanotechnology' is not nanotechnology in the original meaning of the word. Nanotechnology, in its traditional sense, means building things from the bottom up, with atomic precision. This theoretical capability was envisioned as early as 1959 by the renowned physicist Richard Feynman.
I want to build a billion tiny factories, models of each other, which are manufacturing simultaneously. . .The principles of physics, as far as I can see, do not speak against the possibility of maneuvering things atom by atom. It is not an attempt to violate any laws; it is something, in principle, that can be done; but in practice, it has not been done because we are too big. Richard Feynman, Nobel Prize winner in physicsBased on Feynman's vision of miniature factories using nanomachines to build complex products, advanced nanotechnology (sometimes referred to as molecular manufacturing
will make use of positionally-controlled mechanochemistry guided by molecular machine systems. Formulating a roadmap for development of this kind of nanotechnology is now an objective of a broadly based technology roadmap project led by Battelle (the manager of several U.S. National Laboratories) and the Foresight Nanotech Institute.
Shortly after this envisioned molecular machinery is created, it will result in a manufacturing revolution, probably causing severe disruption. It also has serious economic, social, environmental, and military implications.
Four Generations
Mihail (Mike) Roco of the U.S. National Nanotechnology Initiative has described four generations of nanotechnology development (see chart below). The current era, as Roco depicts it, is that of passive nanostructures, materials designed to perform one task. The second phase, which we are just entering, introduces active nanostructures for multitasking; for example, actuators, drug delivery devices, and sensors. The third generation is expected to begin emerging around 2010 and will feature nanosystems with thousands of interacting components. A few years after that, the first integrated nanosystems, functioning (according to Roco) much like a mammalian cell with hierarchical systems within systems, are expected to be developed.
By
CNu
at
October 18, 2011
0
comments
![]()
Labels: microcosmos , Possibilities
Thursday, April 21, 2011
neuroscience of the gut
 Scientific American | People may advise you to listen to your gut instincts: now research suggests that your gut may have more impact on your thoughts than you ever realized. Scientists from the Karolinska Institute in Sweden and the Genome Institute of Singapore led by Sven Pettersson recently reported in the Proceedings of the National Academy of Sciences that normal gut flora, the bacteria that inhabit our intestines, have a significant impact on brain development and subsequent adult behavior.
Scientific American | People may advise you to listen to your gut instincts: now research suggests that your gut may have more impact on your thoughts than you ever realized. Scientists from the Karolinska Institute in Sweden and the Genome Institute of Singapore led by Sven Pettersson recently reported in the Proceedings of the National Academy of Sciences that normal gut flora, the bacteria that inhabit our intestines, have a significant impact on brain development and subsequent adult behavior.We human beings may think of ourselves as a highly evolved species of conscious individuals, but we are all far less human than most of us appreciate. Scientists have long recognized that the bacterial cells inhabiting our skin and gut outnumber human cells by ten-to-one. Indeed, Princeton University scientist Bonnie Bassler compared the approximately 30,000 human genes found in the average human to the more than 3 million bacterial genes inhabiting us, concluding that we are at most one percent human. We are only beginning to understand the sort of impact our bacterial passengers have on our daily lives.
Moreover, these bacteria have been implicated in the development of neurological and behavioral disorders. For example, gut bacteria may have an influence on the body’s use of vitamin B6, which in turn has profound effects on the health of nerve and muscle cells. They modulate immune tolerance and, because of this, they may have an influence on autoimmune diseases, such as multiple sclerosis. They have been shown to influence anxiety-related behavior, although there is controversy regarding whether gut bacteria exacerbate or ameliorate stress related anxiety responses. In autism and other pervasive developmental disorders, there are reports that the specific bacterial species present in the gut are altered and that gastrointestinal problems exacerbate behavioral symptoms. A newly developed biochemical test for autism is based, in part, upon the end products of bacterial metabolism.
But this new study is the first to extensively evaluate the influence of gut bacteria on the biochemistry and development of the brain. The scientists raised mice lacking normal gut microflora, then compared their behavior, brain chemistry and brain development to mice having normal gut bacteria. The microbe-free animals were more active and, in specific behavioral tests, were less anxious than microbe-colonized mice. In one test of anxiety, animals were given the choice of staying in the relative safety of a dark box, or of venturing into a lighted box. Bacteria-free animals spent significantly more time in the light box than their bacterially colonized littermates. Similarly, in another test of anxiety, animals were given the choice of venturing out on an elevated and unprotected bar to explore their environment, or remain in the relative safety of a similar bar protected by enclosing walls. Once again, the microbe-free animals proved themselves bolder than their colonized kin.
Pettersson’s team next asked whether the influence of gut microbes on the brain was reversible and, since the gut is colonized by microbes soon after birth, whether there was evidence that gut microbes influenced the development of the brain. They found that colonizing an adult germ-free animal with normal gut bacteria had no effect on their behavior. However, if germ free animals were colonized early in life, these effects could be reversed. This suggests that there is a critical period in the development of the brain when the bacteria are influential. Fist tap Dorcas Daddy.
By
CNu
at
April 21, 2011
0
comments
![]()
Labels: agency , microcosmos
bacteria divide people into types
 NYTimes | In the early 1900s, scientists discovered that each person belonged to one of four blood types. Now they have discovered a new way to classify humanity: by bacteria. Each human being is host to thousands of different species of microbes. Yet a group of scientists now report just three distinct ecosystems in the guts of people they have studied.
NYTimes | In the early 1900s, scientists discovered that each person belonged to one of four blood types. Now they have discovered a new way to classify humanity: by bacteria. Each human being is host to thousands of different species of microbes. Yet a group of scientists now report just three distinct ecosystems in the guts of people they have studied.Blood type, meet bug type.
“It’s an important advance,” said Rob Knight, a biologist at the University of Colorado, who was not involved in the research. “It’s the first indication that human gut ecosystems may fall into distinct types.”
The researchers, led by Peer Bork of the European Molecular Biology Laboratory in Heidelberg, Germany, found no link between what they called enterotypes and the ethnic background of the European, American and Japanese subjects they studied.
Nor could they find a connection to sex, weight, health or age. They are now exploring other explanations. One possibility is that the guts, or intestines, of infants are randomly colonized by different pioneering species of microbes.
The microbes alter the gut so that only certain species can follow them.
Whatever the cause of the different enterotypes, they may end up having discrete effects on people’s health. Gut microbes aid in food digestion and synthesize vitamins, using enzymes our own cells cannot make.
Dr. Bork and his colleagues have found that each of the types makes a unique balance of these enzymes. Enterotype 1 produces more enzymes for making vitamin B7 (also known as biotin), for example, and Enterotype 2 more enzymes for vitamin B1 (thiamine).
The discovery of the blood types A, B, AB and O had a major effect on how doctors practice medicine. They could limit the chances that a patient’s body would reject a blood transfusion by making sure the donated blood was of a matching type. The discovery of enterotypes could someday lead to medical applications of its own, but they would be far down the road.
“Some things are pretty obvious already,” Dr. Bork said. Doctors might be able to tailor diets or drug prescriptions to suit people’s enterotypes, for example.
Or, he speculated, doctors might be able to use enterotypes to find alternatives to antibiotics, which are becoming increasingly ineffective. Instead of trying to wipe out disease-causing bacteria that have disrupted the ecological balance of the gut, they could try to provide reinforcements for the good bacteria. “You’d try to restore the type you had before,” he said.
Dr. Bork notes that more testing is necessary. Researchers will need to search for enterotypes in people from African, Chinese and other ethnic origins. He also notes that so far, all the subjects come from industrial nations, and thus eat similar foods. “This is a shortcoming,” he said. “We don’t have remote villages.”
The discovery of enterotypes follows on years of work mapping the diversity of microbes in the human body — the human microbiome, as it is known. The difficulty of the task has been staggering. Each person shelters about 100 trillion microbes.
(For comparison, the human body is made up of only around 10 trillion cells.) But scientists cannot rear a vast majority of these bacteria in their labs to identify them and learn their characteristics.
By
CNu
at
April 21, 2011
2
comments
![]()
Labels: microcosmos , What IT DO Shawty...
Self-Proclaimed Zionist Biden Joins The Great Pretending...,
Biden, at today's Holocaust Remembrance Ceremony, denounces the "anti-Semitic" student protests in his strongest terms yet. He...
-
theatlantic | The Ku Klux Klan, Ronald Reagan, and, for most of its history, the NRA all worked to control guns. The Founding Fathers...
-
Video - John Marco Allegro in an interview with Van Kooten & De Bie. TSMATC | Describing the growth of the mushroom ( boletos), P...
-
Farmer Scrub | We've just completed one full year of weighing and recording everything we harvest from the yard. I've uploaded a s...